Speed up a Python program with Rust
In this entry, I showcase how to use Rust to do the legwork in a Python program. I focus on the calculation of Julia sets representations, as this problem provides plenty of eye-candy to lure the innocent reader.
Julia sets
I focus here on the filled Julia set associated with the following complex quadratic polynomial:
$$f(z) = z^2 + c_0$$
where $z$ is a complex number and $c_0$ a fixed complex parameter. The filled Julia set associated with $c_0$ is the set of $z_0$ such as the sequence
$$z_{n+1} = f(z_n) = z_n^2 + c_0$$
does not diverge. A common way to represent those sets is to apply the sequence a few tens or hundreds of times to a region of the complex plane and map how many iterations it takes for the sequence to diverge beyond a chosen magnitude. This is what it looks like for $c_0 = -0.835 -0.2321i$:
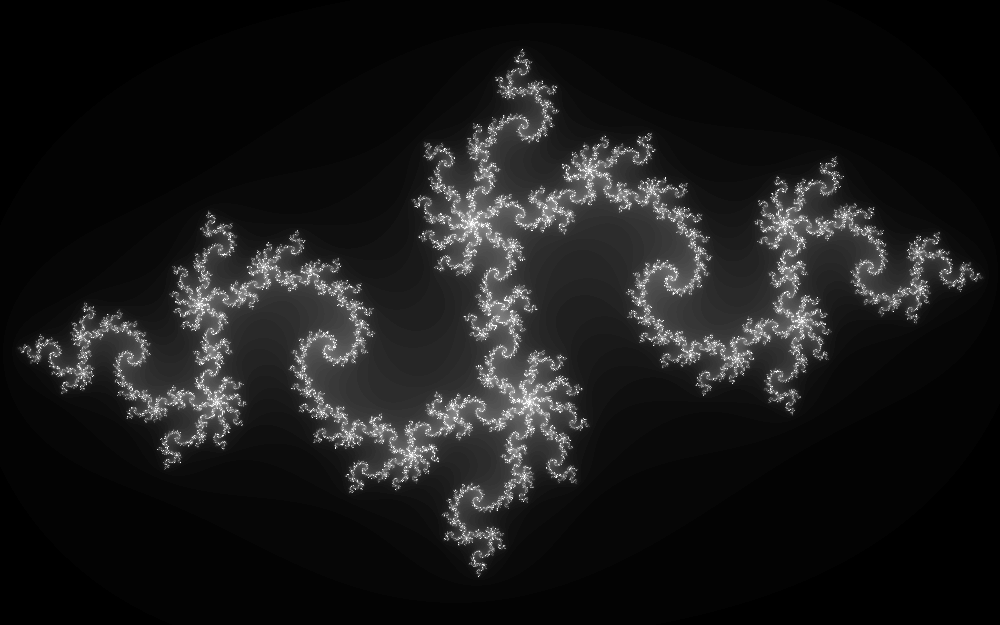
This image has been generated as follow:
- $x$ goes from -1.6 (left) to 1.6 (right), with a resolution of 1000 pixels;
- $y$ goes from -1 (bottom) to 1 (top), with a resolution of 625 pixels;
- each pixel in the image corresponds to a $z_0=x+iy$;
- up to 80 iterations of $z_{n+1} = z_n^2 + c_0$ are performed on each pixel;
- the grayscale represents how many iterations it took to reach $|z_n|\geqslant 2$, going from black (diverged in 1 iteration), to white (has not diverged after 80 iterations).
The image obtained with this procedure is an approximative representation of the filled Julia set, the grayscale giving an idea of how close we are to the set in a particular pixel.
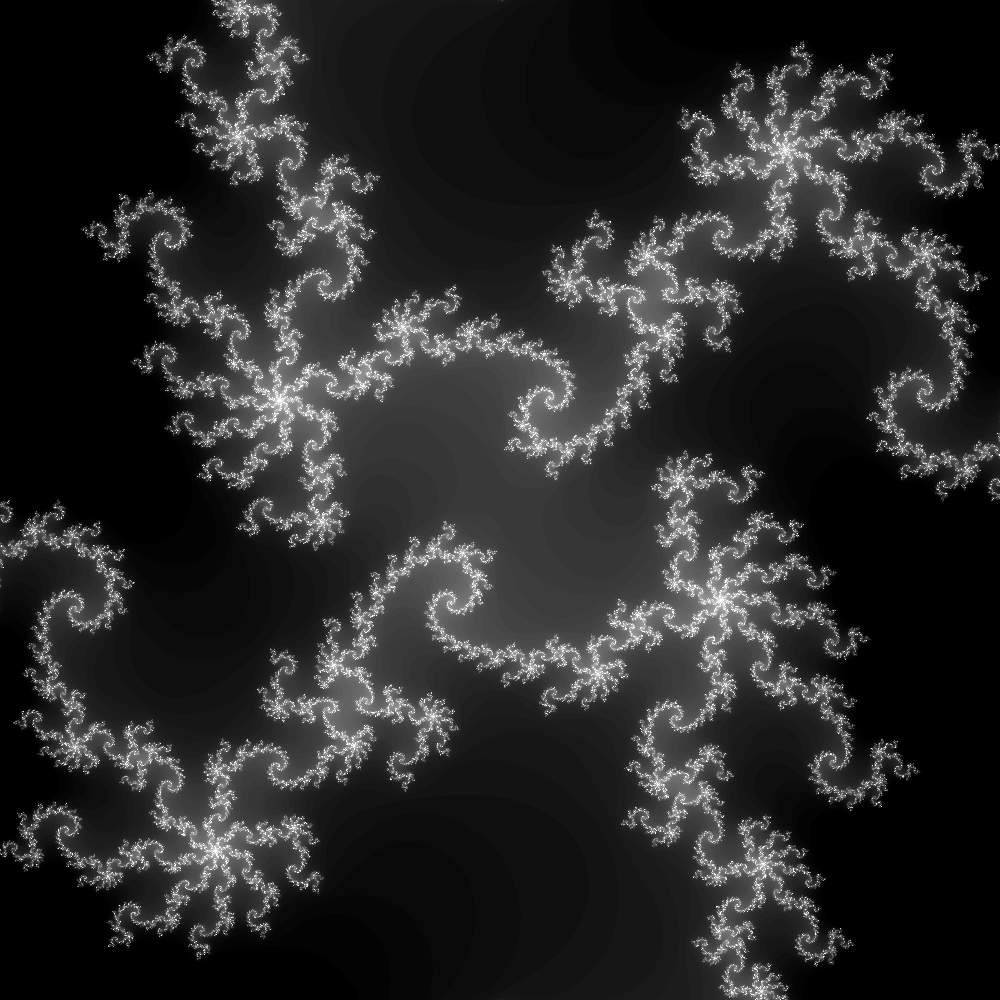
In the rest of this entry, I focus on a smaller region with many details, so that the computation is more demanding. The region is $[-0.1, 0.1]^2$, resolved with 100 iterations and 4000 pixels along each direction. For aesthetic reasons, pixels that have diverged in 20 iterations or fewer are fully black. The full image is accessible here. Here is a smaller resolution image of the same region:
Python implementation with numpy.vectorize
Here is a simple Python implementation. All the
interesting work happens in the divergence function:
def divergence(z_0: complex, c_0: complex, threshold: float, itermax: int) -> int:
for i in range(itermax):
z_0 = z_0**2 + c_0
if abs(z_0) >= threshold:
return i
return itermax
This is a direct implementation of the algorithm described in the previous
section, taking one value of $z_0$ and returning how many iterations it took
for the associated sequence to diverge. This function is then applied to an
array containing all the values of $z_0$ for our image, leveraging
numpy.vectorize:
class JuliaDiv:
# ...
def over(self, plane: ComplexRegion) -> NDArray[np.floating]:
itermin, itermax = self.n_iterations
assert itermin < itermax
z_s = plane.build(self.resolution)
div_vect = np.vectorize(divergence, otypes=[np.uint32])
div = div_vect(z_s, self.c_0, self.threshold, itermax)
return (np.maximum(div, itermin) - itermin) / (itermax - itermin)
In this snippet, z_s is the array containing all the values of z_0 in the
image, div_vect is the vectorized version of divergence, and div is the
array of the number of iterations it took for the corresponding z_0 to
diverge. This number of iterations is then projected on $[0, 1]$ for plotting
purposes. Profiling this version of the code when calculating the target image
leads to the following:
$ python -m cProfile --sort cumulative julia_vectorize.py
544664132 function calls (544660624 primitive calls) in 139.448 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
164/1 0.001 0.000 139.448 139.448 {built-in method builtins.exec}
1 0.001 0.001 139.448 139.448 julia_vectorize.py:1(<module>)
1 0.019 0.019 139.311 139.311 julia_vectorize.py:58(main)
1 0.046 0.046 139.201 139.201 julia_vectorize.py:43(over)
1 0.134 0.134 139.106 139.106 function_base.py:2300(__call__)
1 2.790 2.790 138.972 138.972 function_base.py:2399(_vectorize_call)
16000000 98.280 0.000 135.577 0.000 julia_vectorize.py:28(divergence)
528530089 37.298 0.000 37.298 0.000 {built-in method builtins.abs}
9 0.606 0.067 0.606 0.067 {built-in method numpy.asanyarray}
... and a bunch of function calls with negligible cost
Computing and saving the image takes 2 minutes and 19 seconds, the vast
majority of that time being spent calling and executing divergence. This
already exposes a limitation of the approach in this first implementation:
numpy.vectorize doesn't perform any kind of optimization and merely loops
over the image, calling the Python function on every pixel in a scalar fashion.
This is convenient to call an arbitrary scalar
function over a fairly small array, but costly when dealing with larger arrays.
Python implementation with numpy operations
Can we do better than numpy.vectorize by relying on direct operations on
arrays? Yes, we can! Instead of defining a divergence function and
"vectorize" it, an equivalent computation can be expressed purely with numpy
operations, like so:
class JuliaDiv:
# ...
def over(self, plane: ComplexRegion) -> NDArray[np.floating]:
itermin, itermax = self.n_iterations
assert itermin < itermax
z_s = plane.build(self.resolution)
div = np.full_like(z_s, itermax, dtype=np.uint32)
for i in range(itermax):
z_s = np.square(z_s) + self.c_0
diverged = np.abs(z_s) >= self.threshold
div[diverged] = np.uint32(i)
z_s[diverged] = np.nan
return (np.maximum(div, itermin) - itermin) / (itermax - itermin)
For convenience, the full code with this modification can be accessed
here. This time, the entire z_s array is processed at each
iteration. The points of z_s that have diverged are filled with np.nan,
and the latest points that have diverged are saved in the div array to build
the output image.
Compared to the previous implementation, this is quite wasteful in terms of
calculations as it is unnecessary to keep computing the sequence on points
that have already diverged. However, the computations and looping effectively
happen on the numpy side, i.e. C and Fortran (compiled to machine code)
instead of Python. Let's see the profile:
$ python -m cProfile --sort cumulative julia.py
134133 function calls (130623 primitive calls) in 19.057 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
164/1 0.001 0.000 19.057 19.057 {built-in method builtins.exec}
1 0.000 0.000 19.057 19.057 julia.py:1(<module>)
1 0.020 0.020 18.920 18.920 julia.py:48(main)
1 18.802 18.802 18.858 18.858 julia.py:35(over)
17 0.001 0.000 0.262 0.015 __init__.py:1(<module>)
... and a bunch of function calls with negligible cost
This change already shaves off 2 minutes of the 2 minutes and 19 seconds, leading to a 7-fold speedup. In this case, it is much more performant to call operations that are vectorized at the machine code level even if it means doing some extraneous calculations.
Python implementation with Numba
Numba is a JIT compiler, interacting particularly
well with numpy. In particular, it widens the set of operations that can
be efficiently vectorized over numpy arrays by compiling supported Python
operations to machine code. In the present case, our function divergence is
simple enough that numba.vectorize can act as a drop-in replacement of
numpy.vectorize for a huge speedup:
@numba.vectorize(
["uint32(complex128, complex128, float64, uint32)"],
)
def divergence(z_0: complex, c_0: complex, threshold: float, itermax: int) -> int:
for i in range(itermax):
z_0 = z_0**2 + c_0
if abs(z_0) >= threshold:
return i
return itermax
With this simple modification, the runtime is down to 6 seconds! This is already an impressive 3-fold speedup from the previous implementation. Let's see what our program spends its time doing:
$ perf record python julia_numba.py
$ perf report
51.26% python libm.so.6 [.] hypot
7.83% python [JIT] tid 3393 [.] 00007ff03763d2a2
4.08% python [JIT] tid 3393 [.] 0x00007ff03763d2a9
2.27% python [JIT] tid 3393 [.] 0x00007ff03763d270
2.24% python [JIT] tid 3393 [.] 0x00007ff03763d29c
2.13% python [JIT] tid 3393 [.] 0x00007ff03763d297
2.10% python [JIT] tid 3393 [.] 0x00007ff03763d2b5
2.01% python [JIT] tid 3393 [.] 0x00007ff03763d283
1.92% python _imaging.cpython-310-x86_64-linux-gnu.so [.] ImagingZipEncode
1.70% python libpython3.10.so.1.0 [.] _PyEval_EvalFrameDefault
0.82% python libz.so.1.2.12 [.] 0x00000000000033b5
...
Half the runtime is spent in libm::hypot, i.e. computing
$|z_n|=\sqrt{x^2+y^2}$. This seems like a good time to sacrifice a little bit
of readability to try and cut down this time:
@numba.vectorize(
["uint32(complex128, complex128, float64, uint32)"],
)
def divergence(z_0: complex, c_0: complex, threshold: float, itermax: int) -> int:
thr_sqr = threshold**2
for i in range(itermax):
z_0 = z_0**2 + c_0
if z_0.real**2 + z_0.imag**2 >= thr_sqr:
return i
return itermax
We're now down to a runtime of 3.2 seconds! The full code is here.
Numba is an excellent tool to speed up a Python program that manipulates numpy arrays with minimal efforts. For the problem at hand, this is a very reasonable solution. However, it is limited to the operations Numba can compile to efficient machine code. It can also be challenging to profile the application and assess how to further improve performances if need be.
Rust-backed implementation
A radical and very effective way to speed up a Python application is to rewrite the most demanding parts in a language that compiles to fast machine code and expose that code to Python via the Python C API. This is essentially how numpy manages to be so fast: it is coded mainly in C and Fortran, and exposes a Python API that the end-user calls. A combo that I find particularly ergonomic to do this is the Rust programming language to write the demanding parts, along with the PyO3 project, a collection of Rust libraries (a.k.a. crates) to bridge Rust and Python.
In this section I show how to write a Rust-powered library that is a drop-in
replacement of the previous Python implementation of the ComplexRegion and
JuliaDiv classes. Here is the full implementation, that you can explore as you follow along
the explanations. Note that the code is split in two crates (leveraging
the workspace feature of Cargo):
- the Rust implementation itself, a pure Rust library at the root of the repository;
- the library exposing a Python API in the
/py-apifolder.
One could choose a more direct organisation with only one library exposing a Python API and keep the pure Rust library as a private implementation detail. However, separating the Rust library and the Python API has multiple advantages:
- the pure Rust library can be easily tested on its own, without Python-related considerations;
- Rust projects can use the Rust library without caring about the Python side of things at all;
- it is much easier to keep the layer between Rust and Python as thin as possible, avoiding e.g. GIL-related issues as much as possible.
Please note that the goal of these explanations is not to be a viable substitute for PyO3 documentation. Duplicating the latter would be pointless and doomed to be out-of-date sooner or later. This article only aims at providing broad explanations on how to organize a project to use PyO3 and introducing some of its features, in the hope that it piques your interest.
Pure Rust library
The implementation (in /src/lib.rs) is fairly straightforward. It is
voluntarily coded in a very similar fashion to the Python code used until now.
It uses ndarray to manipulate 2D
arrays, and num-complex to
manipulate complex numbers.
[package]
name = "juliaset"
version = "0.1.0"
edition = "2021"
rust-version = "1.60"
[dependencies]
ndarray = "0.15.6"
num-complex = "0.4.2"
The core of the computation happens in the JuliaDiv::over method:
impl JuliaDiv {
pub fn over(&self, plane: &ComplexRegion) -> Array2<f64> {
let (itermin, itermax) = self.n_iterations;
let thres_sqr = self.threshold.powi(2);
plane.build(self.resolution).mapv_into_any(|mut z_s| {
let mut i = 0;
loop {
z_s = z_s * z_s + self.c_0;
if i == itermax || z_s.norm_sqr() > thres_sqr {
break;
}
i += 1;
}
(i.max(itermin) - itermin) as f64 / (itermax - itermin) as f64
})
}
}
In particular, ArrayBase::mapv_into_any offers a very ergonomic way to map each $z_0$ to the "divergence number".
Exposing Rust code as a Python API
Once the core Rust library is implemented, the next step is to build a thin
layer around its public API to expose it to Python. As discussed earlier, this
is done in a separate crate in the same workspace to keep the interface between
Rust and Python nice and tidy. Adding a new crate in the current workspace is
as simple as adding two lines in the main /Cargo.toml:
[package]
name = "juliaset"
version = "0.1.0"
edition = "2021"
rust-version = "1.60"
[dependencies]
ndarray = "0.15.6"
num-complex = "0.4.2"
[workspace]
members = ["py-api"]
where each element of the members list is a directory containing a crate.
Its own manifest /py-api/Cargo.toml is fairly standard:
[package]
name = "juliaset-py"
version = "0.1.0"
edition = "2021"
rust-version = "1.60"
[lib]
name = "juliaset"
crate-type = ["cdylib"]
[dependencies]
juliaset = { path = "..", version = "0.1.0" }
numpy = "0.17.1"
pyo3 = { version = "0.17.1", features = ["extension-module", "abi3-py37", "num-complex"] }
Three lines, highlighted above, are noteworthy:
- I ask Rust to build a
"cdylib", i.e. a regular shared library (a.soon Linux) rather than a Rust library as this is what Python can understand and load; - the
juliasetpure Rust library that we wrote before is marked as a dependency; - the following features of PyO3 are used:
extension-moduleto use the tools necessary to expose Rust code as a Python API;num-complexto handle complex types and allow automatic conversions between Python types andComplex64on Rust side;abi3-py37to build wheels that are compatible with Python 3.7 and above rather than only a specific Python version.
A straighforward strategy to write the Python API is to wrap the underlying
Rust types in a newtype that is a pyclass (i.e. that Python can see as a
regular class). For instance, the Python counterpart to
juliaset::ComplexRegion can be declared as follows:
use pyo3::prelude::*;
/// Define an area of the complex plane.
#[pyclass(frozen)]
pub struct ComplexRegion(::juliaset::ComplexRegion);
All the magic happens thanks to the #[pyclass] attribute. PyO3 implements the
necessary machinery to expose ComplexRegion as a Python class. It is marked
as frozen here to make the instances immutable. As a nice perk, the docstring
of the Python class has the same content as the doc-comment.
We then need to define methods, which can be done with an impl block with the
#[pymethods] attribute:
use numpy::{Complex64, PyArray2};
#[pymethods]
impl ComplexRegion {
#[new]
fn new(xleft: f64, xright: f64, yleft: f64, yright: f64) -> Self {
Self(::juliaset::ComplexRegion::new(xleft, xright, yleft, yright))
}
/// Create an array spanning the region with the given resolution along the real axis.
fn build<'py>(&self, py: Python<'py>, resolution: usize) -> &'py PyArray2<Complex64> {
let out = self.0.build(resolution);
PyArray2::from_owned_array(py, out)
}
}
The #[new] attribute marks the constructor method. This one is
straightforward, PyO3 even handles the float to f64 conversion for us.
The build method is a little more interesting. This method returns a numpy
array (numpy::PyArray on Rust's side). The lifetime of the latter is managed
at runtime by Python rather than statically by Rust's borrow checker, which is
why we need to take a Python<'py> token. This token doesn't appear in the
method signature when calling it from Python, it is merely the way PyO3 models
holding the Global Interpreter Lock of Python. Making the widely different
memory management models of Rust and Python play nicely together is not a
trivial task, but thankfully PyO3 does a great job at abstracting that problem
for us. As you can see in this example, the numpy crate offers a
straighforward way to transform the owned ndarray::Array we get from
juliaset::ComplexRegion::build into the Python-managed numpy::PyArray we
want.
The final step needed to expose our class to Python is to add it to a Python module. Once again, PyO3 makes it fairly straighforward:
/// Routines to compute Julia sets for imaging purposes
#[pymodule]
fn juliaset(_py: Python<'_>, pymod: &PyModule) -> PyResult<()> {
pymod.add_class::<ComplexRegion>()?;
Ok(())
}
One noteworthy detail here is the PyResult<T> return type. It is an alias
of Result<T, PyErr>, and more importantly is the way PyO3 models operations
that can raise Python exceptions. As adding a class to a module is a faillible
operation, it returns a PyResult. If the operation fails, it returns a
Result::Err(PyErr) and you end up with an exception on Python side.
Building and using the Python package
As shown previously, PyO3 makes it easy to expose Rust code through a Python
API. It also makes it easy to build a Python wheel using
maturin. All that is needed to build and install our
crate as a Python package is the following /py-api/pyproject.toml:
[build-system]
requires = ["maturin>=0.13.2,<0.14"]
build-backend = "maturin"
[project]
name = "juliaset"
requires-python = ">=3.7"
dependencies = [
"numpy>=1.21",
]
The highlighted lines declare maturin as build system for our Python package,
and numpy as a runtime dependency (required by the numpy crate on Rust's
side). Installing our crate as a Python package is then as simple as
$ python3 -m pip install ./py-api
We can then import and use it as any other Python package, here is a Python
program that uses our package to produce the target image.
The runtime is now down to 2.8s, a further ~12% improvement from the 3.2s with
numba! While the performance reached with numba is certainly impressive
and probably sufficient in plenty of contexts, this shows a bespoke low-level
code can still be significantly faster and worth the effort.
The PyO3 project offers ways to build and publish wheels for several platforms, please see the documentations for more information.
One thing still missing from PyO3 at the time of writing is the automatic
generation of type annotations of the Python library, e.g. for consumption by
mypy (static type checker) or a
language server.
This requires manually writing a stub with type annotations if you care about
these things, see /py-api/juliaset.pyi for our package.
Rust CLI
For comparison purposes, I also wrote a CLI in Rust around the pure Rust
library. It is behind the cli feature to avoid having unused dependencies
when building only the library. The relevant parts of /Cargo.toml are
highlighted here:
[package]
name = "juliaset"
version = "0.1.0"
edition = "2021"
rust-version = "1.60"
[dependencies]
clap = { version = "4.0.1", features = ["derive"], optional = true }
image = { version = "0.24.3", default-features = false, features = ["png"], optional = true }
ndarray = "0.15.6"
num-complex = "0.4.2"
[features]
cli = ["dep:image", "dep:clap"]
[[bin]]
name = "juliaset"
required-features = ["cli"]
[workspace]
members = ["py-api"]
clap manages command line arguments, and image allows us to easily create a
picture from an array.
$ cargo run --release -F cli -- -h
builds and run the CLI tool, asking it to display its help message (-h
option) generated by clap.
It has a similar runtime to the Python program calling the Rust backend (2.8s).
$ time ./target/release/juliaset -x=-0.1 -X=0.1 -y=-0.1 -Y=0.1 -m=20 -M=100 -R=4000
./target/release/juliaset -x=-0.1 -X=0.1 -y=-0.1 -Y=0.1 -m=20 -M=100 -R=4000 2.83s user 0.05s system 99% cpu 2.876 total
Note however that the obtained image is more compressed than the Python counterpart (2.2 MB vs 2.4 MB). Changing the compression algorithm can lead to huge differences on the runtime as a significant chunk of the runtime is now spent compressing the output image:
$ perf report
69.60% juliaset juliaset [.] ndarray::impl_methods::<impl ndarray::ArrayBase<S,D>>::mapv_into_any
27.48% juliaset juliaset [.] miniz_oxide::deflate::core::compress_inner
1.07% juliaset juliaset [.] ndarray::iterators::to_vec_mapped
0.63% juliaset juliaset [.] ndarray::iterators::to_vec_mapped
0.51% juliaset juliaset [.] ndarray::impl_methods::<impl ndarray::ArrayBase<S,D>>::map
0.33% juliaset juliaset [.] miniz_oxide::deflate::core::compress_block
...
If only minimal compression is used (--fast flag of the CLI), the file
produced takes up 3.1 MB and the runtime falls to 2.1s.
$ perf report # with minimal compression
94.12% juliaset juliaset [.] ndarray::impl_methods::<impl ndarray::ArrayBase<S,D>>::mapv_into_any
1.77% juliaset juliaset [.] miniz_oxide::deflate::core::compress_inner
1.39% juliaset juliaset [.] ndarray::iterators::to_vec_mapped
0.84% juliaset juliaset [.] ndarray::iterators::to_vec_mapped
0.80% juliaset juliaset [.] miniz_oxide::deflate::core::compress_block
0.66% juliaset juliaset [.] ndarray::impl_methods::<impl ndarray::ArrayBase<S,D>>::map
...
We now spend 94% of the runtime computing what we're interested in, that's pretty great!
Closing remarks
I hope this entry convinced you that Rust with PyO3 is a viable candidate to
write computational-intensive parts of Python programs that cannot be easily
expressed with numpy. The following table summarizes the timings of various
implementations:
| Implementation | Runtime (s) | Speedup naive Python | Speedup previous |
|---|---|---|---|
naive Python (np.vectorize) | 139 | 0% | - |
| numpy looping | 19 | 86.3% | 86.3% |
| numba | 6.0 | 95.7% | 68.4% |
| numba (no square root) | 3.2 | 97.7% | 46.7% |
| Rust backend / Rust CLI | 2.8 | 98.0% | 12.5% |
| Rust CLI (minimal compression) | 2.1 | 98.5% | 25.0% |
A general strategy to improve the performances of a Python program that cannot
be quite expressed with numpy loops or methods would be to try the following
implementations:
- wrap in
np.vectorizethe calculation written in Python; - rewrite the computation to use implicit
numpyloops and methods as much as possible, even if it means doing more work; - wrap in
numba.vectorizethe calculation written in Python; - write a bespoke low-level implementation of the desired calculation and expose it to Python.
For most problems, there is a good chance that each of those implementations will yield better performance than the previous one. However, as always when dealing with performances, results may vary widely depending on the hardware, the versions of the tools, the operating system, and, more importantly, the actual problem at hand. In particular, the effective speed-up gained between each of those implementations is highly problem-dependent. Profiling and measuring is in any case the only sure way to identify bottlenecks and which optimizations actually improve performance.